How To Suck At Being A Bioinformatician
Hope you do not learn anything from here
Table of contents
Introduction
While there's plenty of information on how to be a great bioinformatician, there's almost* nothing out there to help you become the opposite – not a bioinformatician. So, here, we've collected some simple rules and a code of behaviour to help those who want to avoid success in bioinformatics. By carefully following these guidelines, you can be assured of going backwards in your bioinformatics career.
With the advent of AI (Artificial Intelligence) and ML (Machine Learning), a large number of Computer Science and Biologists have refocused their attention towards the field of bioinformatics. To add to the challenge, there's the looming possibility of widespread layoffs in the financial sector. Some people are also seeking a new sense of purpose, perhaps triggered by a mid-life crisis, leading to the emergence of a unique breed of bioinformaticians. This diverse group includes:
Those who would rather use their graphics cards for scientific calculations rather than video games and crypto mining, aiming to unlock the secrets of life.
Others who are deeply immersed in the ever-expanding world of -omics subfields (theoretically, but with a great number of irrelevant publications).
Some who primarily work on annotating data are often referred to humorously as "annotation monkeys", with no idea of what to do with the output.
And as if things weren't already complex, there's a constant influx of new software, data formats, and databases in the biomedical field, seemingly adding complexity just for the sake of it. Bioinformatics has become the more affordable alternative to traditional biology, especially during periods of limited funding.
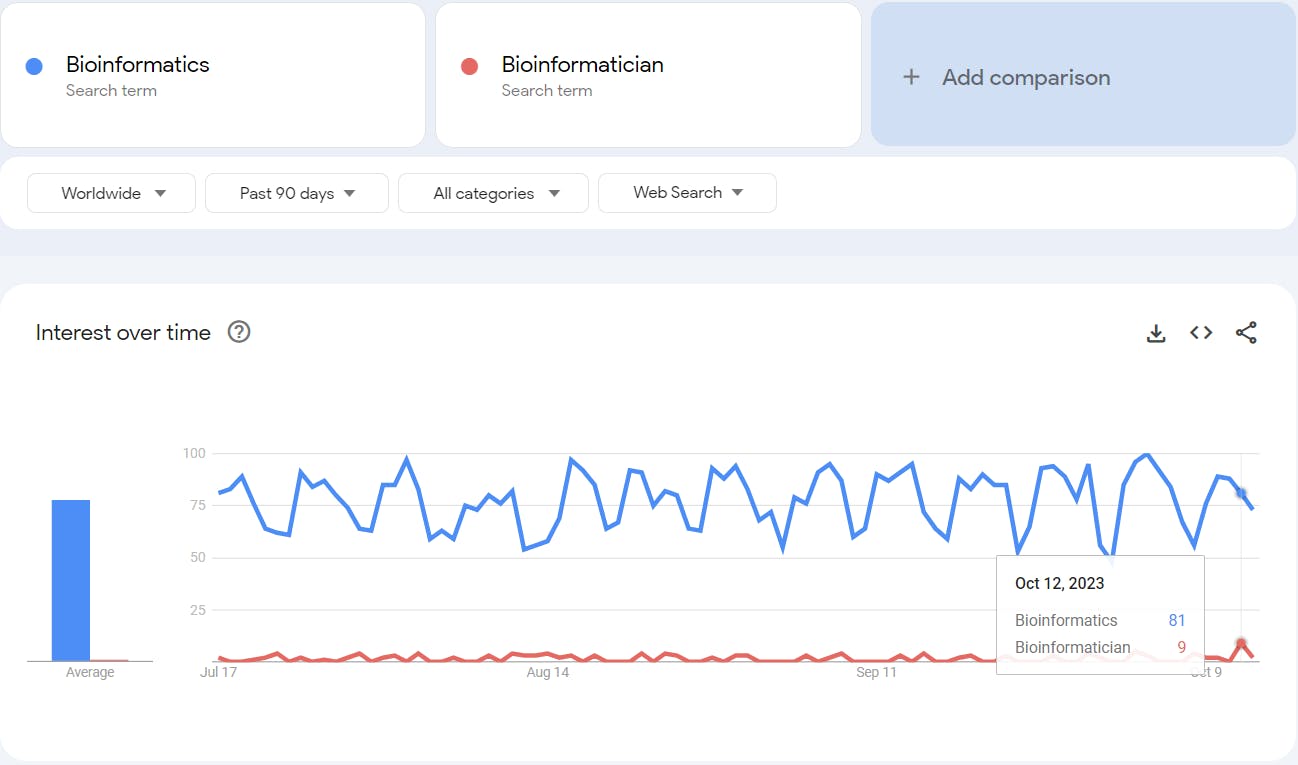
Look for yourself, though not the right comparison, the number of individuals searching to learn about bioinformatics, versus the number of individuals trying to understand the role of bioinformaticians.
Besides the fact that bioinformaticians might seem unnecessary when ChatGPT can provide answers to everything, it's important to note that the suggested approach below, while it has its uses, should not be blindly adhered to:
Methodology
So here are the things you should definitely follow to be a bad Bioinformatician or be bad at bioinformatics (there does exist a difference between both):
Keep it simple and basic at all times: Craft your code as you go, without bothering with planning, requirements, or structured approaches. Don't get caught up in the complexities of object-oriented programming. Instead, create your own collection of small helper scripts. Forget about documenting your code, both internally and externally; let your coding style remain your secret. Ensure your software doesn't handle growth well. Avoid modelling or abstraction and always opt for a quick and straightforward solution.
I absolutely look up to my colleague who chose to spend a day looping over annotated gvcf files instead of using dataframes, for context either indexing or using dataframe would have taken a whopping 20 minutes for the same.
Embrace open source but keep everything cryptic: Error messages are a no-go, but if you absolutely have to include them, make sure they're utterly puzzling. When you build an application, make it as tricky as possible. Throw in lots of hidden dependencies and weird variables. Don't waste time debugging or thinking about compatibility with older versions.
Make your code non-portable, so it only runs on outdated operating systems, and assume that only you will ever use your application. Expect that no one else will understand it. If not all, just host them on a private repository, forget about reproducibilty or validation but do post the titles and output on LinkedIn.
Don't bother with a fancy graphical user interface or APIs: The command line is the way to go and force your end users to use it. Make it extra confusing by using parameter names that have nothing to do with what they actually do. For instance, don't use "-o" for setting an output file; opt for something like "k" or "B" to keep things memorable.
Ensure that the results you share are cryptic – They shouldn't make sense, be impossible to interpret, and definitely not conform to any established standards. Stick with plain ASCII text or, even better, introduce your own scrambled format. Avoid using ontologies, XML, or any other commonly recognized interchange format. If you do happen to use XML, make it as tricky as possible to validate and ensure it doesn't follow any XML schema.
Feel free to make up a new name for a gene, especially if it doesn't match any existing reference database identifier, and keep it to yourself.
Follow industry-drowning best practices: Make sure your Docker containers are bewildering, and impossible to manage, the containerised application should be in the temp/ folder since bin/ is too basic and don't conform to any established standards, even better do not bother to learn what containerisation is as listed in the article below. since what works on your system should work on your system alone.
Stick with obscure configurations, and steer clear of well-documented practices or interoperable standards. When it comes to pipeline frameworks, use loops and bash scripting in the most elaborate way possible, do not use comprehensions or the actual framework itself. Opt for convoluted and unpredictable processes that no one can follow. Create your own unique terminology and processes that leave others scratching their heads.
Don't use established CI/CD methodologies, git is for boomers. Keep your methods and approaches a well-guarded secret.
Put your faith in predictions, P-values, or statistics without question: Cherry-pick data for your training set that guarantees the desired outcome. Set arbitrary cutoffs on ranked results, where the top results are seen as undeniable truth, while everything else is dismissed as false.
Stick to default settings and avoid exploring alternative algorithm options. If you receive a list of results, only pay attention to the very first one. Ignore the "garbage in, garbage out" warning and instead focus on making your flawed data look presentable with colorful plots and vibgyor borders and highlights, spend hours correcting the legend placement.
You're the all-knowing evolved individual: Completely disregard and overlook the guidance provided by an experienced mentor on programming logic during scripting and the significance of biology when implementing that logic.
Definitely don't bother mentioning your implementation. Attend conferences and keep asking the same unimportant questions about the data sequencing platform, all while enjoying a good nap for half of the event.
However, to maintain the illusion, publish a research paper once a year, incorporating the aforementioned practices, just to divert attention.
Conclusion
Here we have highlighted a series of disastrous practices in the bioinformatics field. We hope that readers will not only find them stimulating but useful.
Embrace them fully and we guarantee their efficacy.
P.S. Do not add references to the content like I have below, own it as if you built it.
References:
Bourne P. E. (2021). Is "bioinformatics" dead?. PLoS biology, 19(3), e3001165. https://doi.org/10.1371/journal.pbio.3001165
Corpas, M., Fatumo, S., & Schneider, R. (2012). How not to be a bioinformatician. Source code for biology and medicine, 7(1), 3. https://doi.org/10.1186/1751-0473-7-3
Ouzounis C. A. (2012). Rise and demise of bioinformatics? Promise and progress. PLoS computational biology, 8(4), e1002487. https://doi.org/10.1371/journal.pcbi.1002487